Deep brain - Type 1 - Training
Yanyu Liang
07 May, 2017
1 Overview
Here we train the model on the basis of DeepSEA result, and particularly:
- Use exactly the same sequences for training, validation and testing
- Use the feature representation generated by DeepSEA
- Add new labels, DNase data for E081, E082 in Roadmap, Noonan_H3K27ac (from Yuwen), and H3K9me3 data of E129 in Roadmap as a control, which is one the label in DeepSEA model. The information of Roadmap data can be found here.
- Train a logistic regression classifier
2 Generate feature representation
925-long feature vector for each sequence is computed using script /project2/xinhe/yanyul/deep_variant/yanyu/DeepSEA/my_scripts/run_DeepSEA_keras_feature.py. One of the sbatch script can be found at /project2/xinhe/yanyul/deep_variant/yanyu/deep_brain/my_scripts/train_feature.sbatch. The output data is at:
- Train:
/project2/xinhe/yanyul/deep_variant/yanyu/deep_brain/DeepSEA_train_12_.hdf5 - Valid:
/project2/xinhe/yanyul/deep_variant/yanyu/deep_brain/DeepSEA_valid_12_.hdf5 - Test:
/project2/xinhe/yanyul/deep_variant/yanyu/DeepSEA/my_test/test_all_DeepSEA_keras_12_.hdf5
3 Add new labels
This step is implemented at /project2/xinhe/yanyul/deep_variant/yanyu/deep_brain/my_scripts/wrapper_label_intervals.py. The usage is described in details at here. It also describes how to get the new labels merged with feature representation to get them prepared as input for training script.
4 Train a logistic regression classifier
This part is implemented by Keras, and in principle, we can add other classifiers by specifying the model generation within /project2/xinhe/yanyul/deep_variant/yanyu/deep_brain/my_scripts/deep_brain_my_lib.py. To make things consistent with DeepSEA, we use logistic regression at present.
Another thing to point out is that we use SGD instead of RMSprop since RMSprop converges more slowly in our problem.
We repeat the training five times and one the scripts used is /project2/xinhe/yanyul/deep_variant/yanyu/deep_brain/my_train/040417_merged/logistic_1.sbatch. Every time, we run 10 epochs and the mini-batch size is 100.
The result is generated by the following command:
$ python my_scripts/summary_run.py my_scripts/ my_train/DeepSEA_test_12__with_label.hdf5 E081,E082,E129,Noonan no my_train/040417_overnight/1 my_train/040417_overnight/2 my_train/040417_overnight/3It generates one summary per folder (namely per training). Within each summary file, it contains AUCs, accuracy, precision, recall information computed using test data along the optimization (but loss and val_loss is computed based on train and validation data). The following subsections show the result.
4.1 Load summary files
filenames = 1 : 3
summary_table <- c()
for(i in filenames){
temp <- read.table(paste('../data/040417_overnight/', paste(i, 'csv', sep = '.'), sep = ''), sep = ',', header = T)
temp$repeatID <- rep(i, nrow(temp))
summary_table <- rbind(summary_table, temp)
}4.2 Loss and validation loss
library(ggplot2)
ggplot() + geom_line(data = summary_table[summary_table$variable == 'loss',], aes(x = epoch, y = value, group = repeatID, color = 'loss')) + geom_line(data = summary_table[summary_table$variable == 'val_loss',], aes(x = epoch, y = value, group = repeatID, color = 'val_loss')) + ggtitle('Loss and validation loss along the optimization')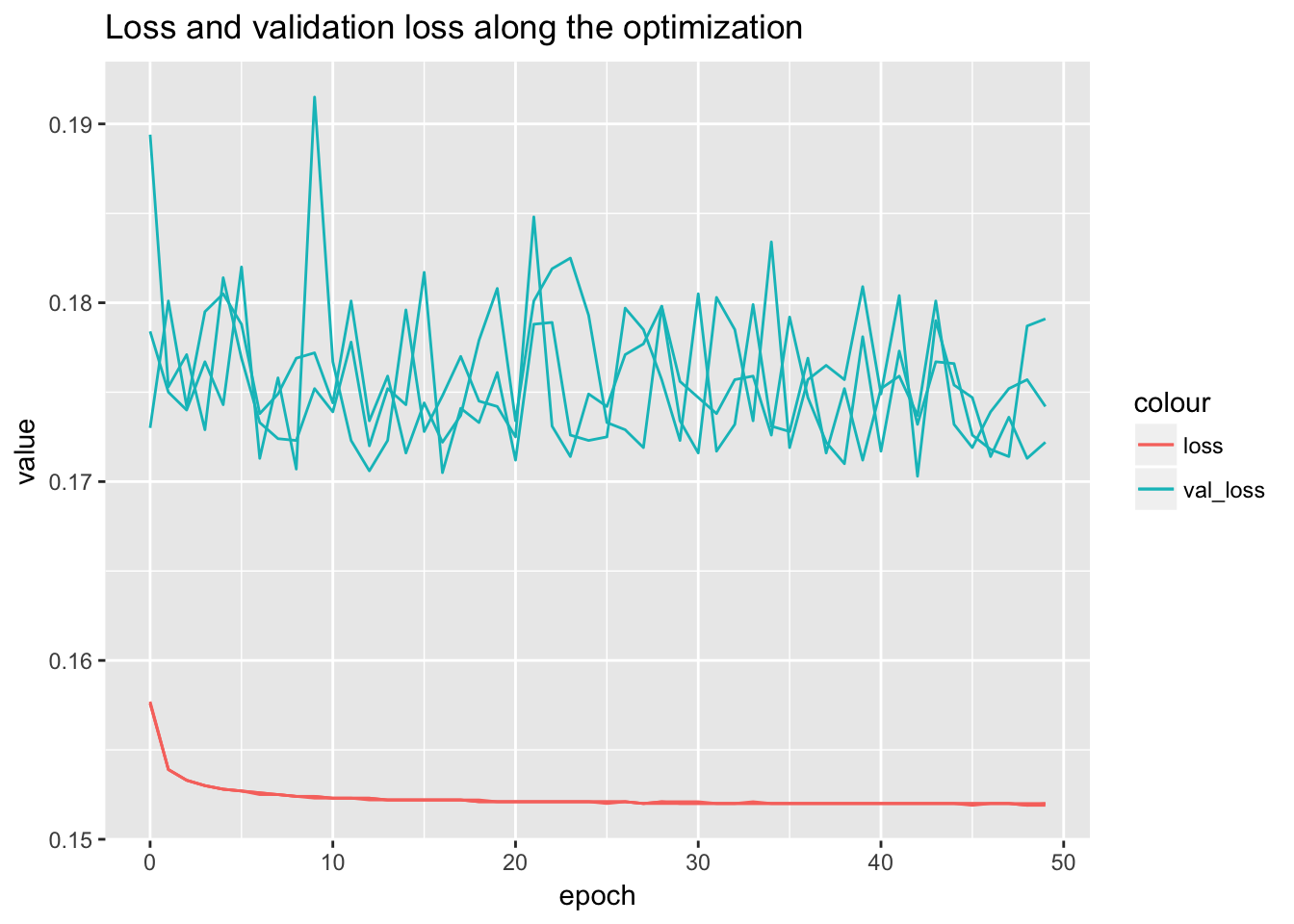
4.3 Precision, recall, and AUCs
temp <- summary_table[!summary_table$variable %in% c('loss', 'val_loss'), ]
temp$type <- factor(temp$variable, levels = c('cross_entropy', 'roc_auc', 'pr_auc', 'accuracy', 'precision_f', 'precision_t', 'recall_f', 'recall_t', 'proportion_f', 'proportion_t'))
ggplot(temp) + geom_boxplot(aes(x = type, y = value, color = data)) + theme(axis.text.x = element_text(angle = 30, hjust = 1)) + ggtitle('Performance overview')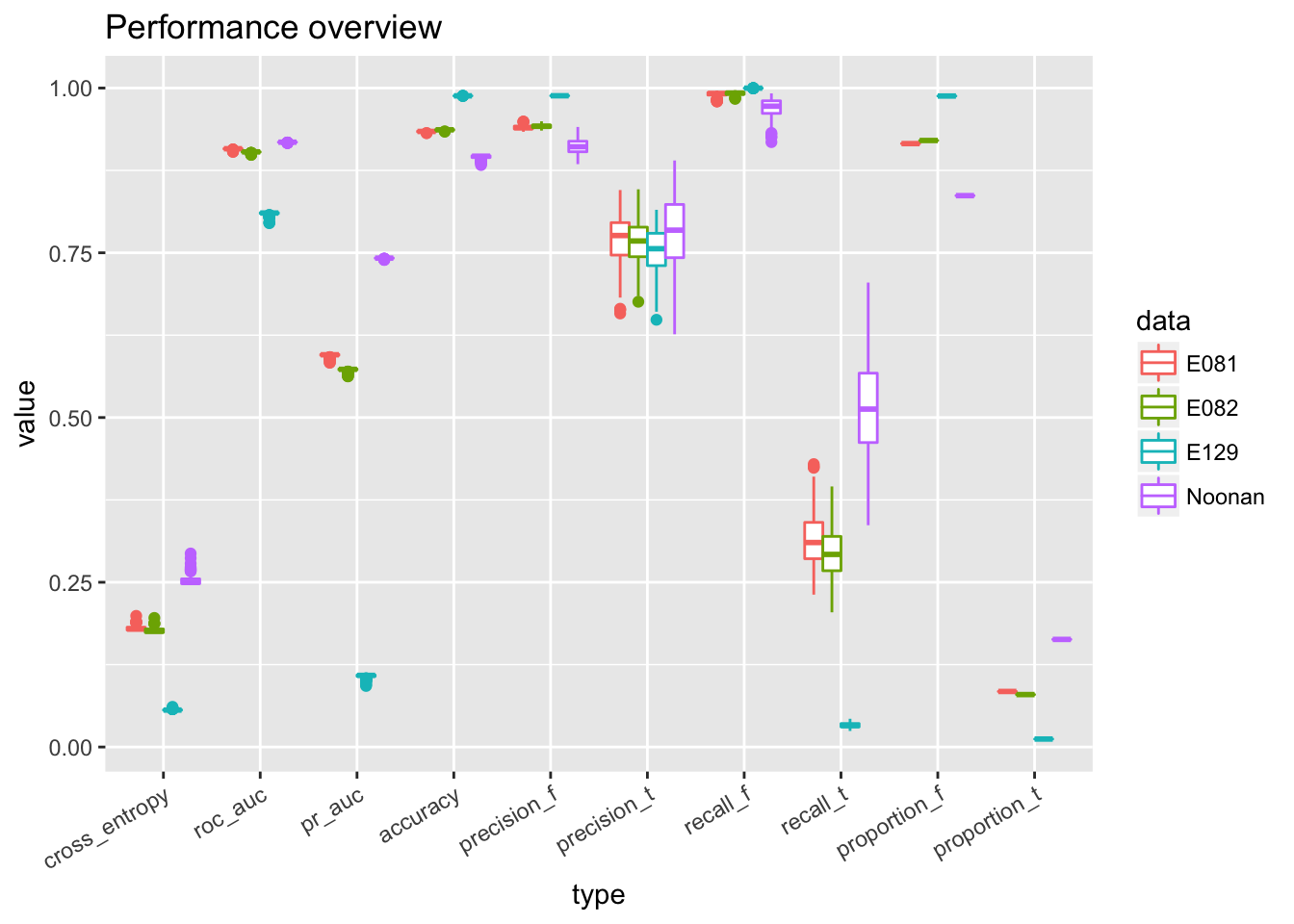
temp.no_proportion <- temp[!grepl('proportion', temp$type), ]
ggplot(temp.no_proportion[temp.no_proportion$data == 'Noonan',]) + geom_line(aes(x = epoch, y = value, group = repeatID)) + facet_grid(type ~ ., scales = 'free_y', ) + ggtitle('Noonan H3K27ac') + theme(strip.text.y = element_text(angle = 0))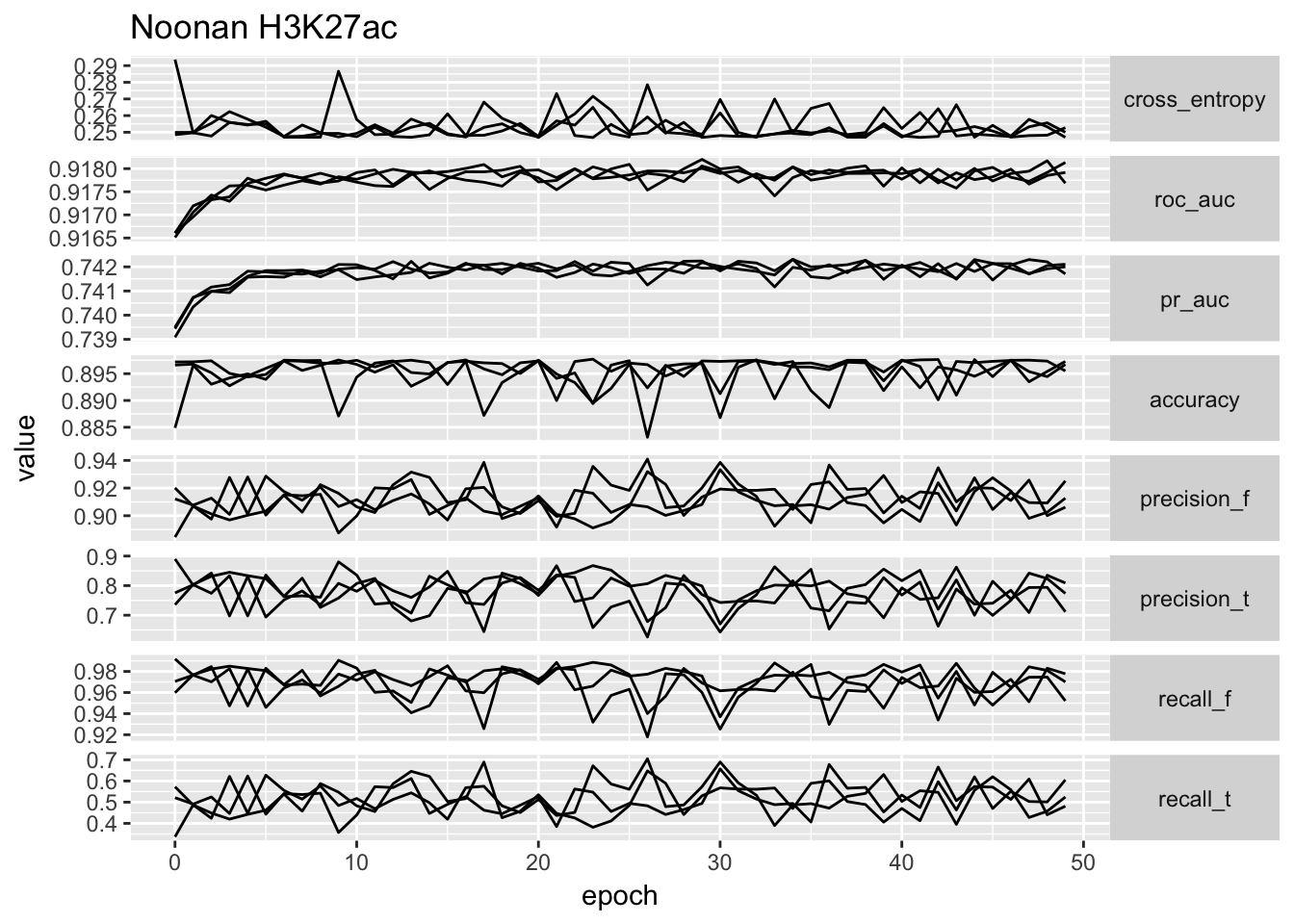
ggplot(temp.no_proportion[temp.no_proportion$data == 'E129',]) + geom_line(aes(x = epoch, y = value, group = repeatID)) + facet_grid(type ~ ., scales = 'free_y', ) + ggtitle('E129 H3K9me3') + theme(strip.text.y = element_text(angle = 0))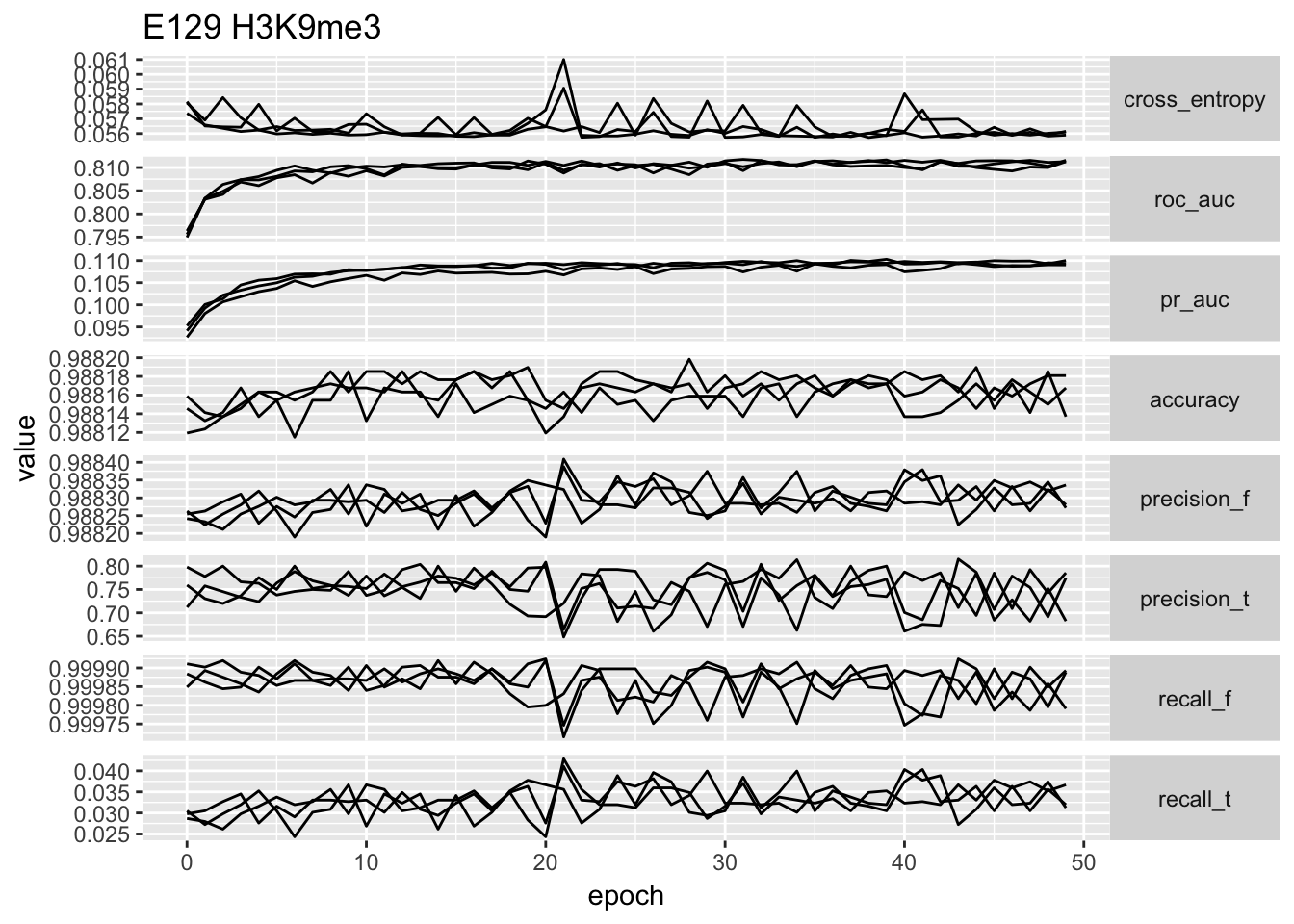
ggplot(temp.no_proportion[temp.no_proportion$data == 'E081',]) + geom_line(aes(x = epoch, y = value, group = repeatID)) + facet_grid(type ~ ., scales = 'free_y', ) + ggtitle('E081 DNase') + theme(strip.text.y = element_text(angle = 0))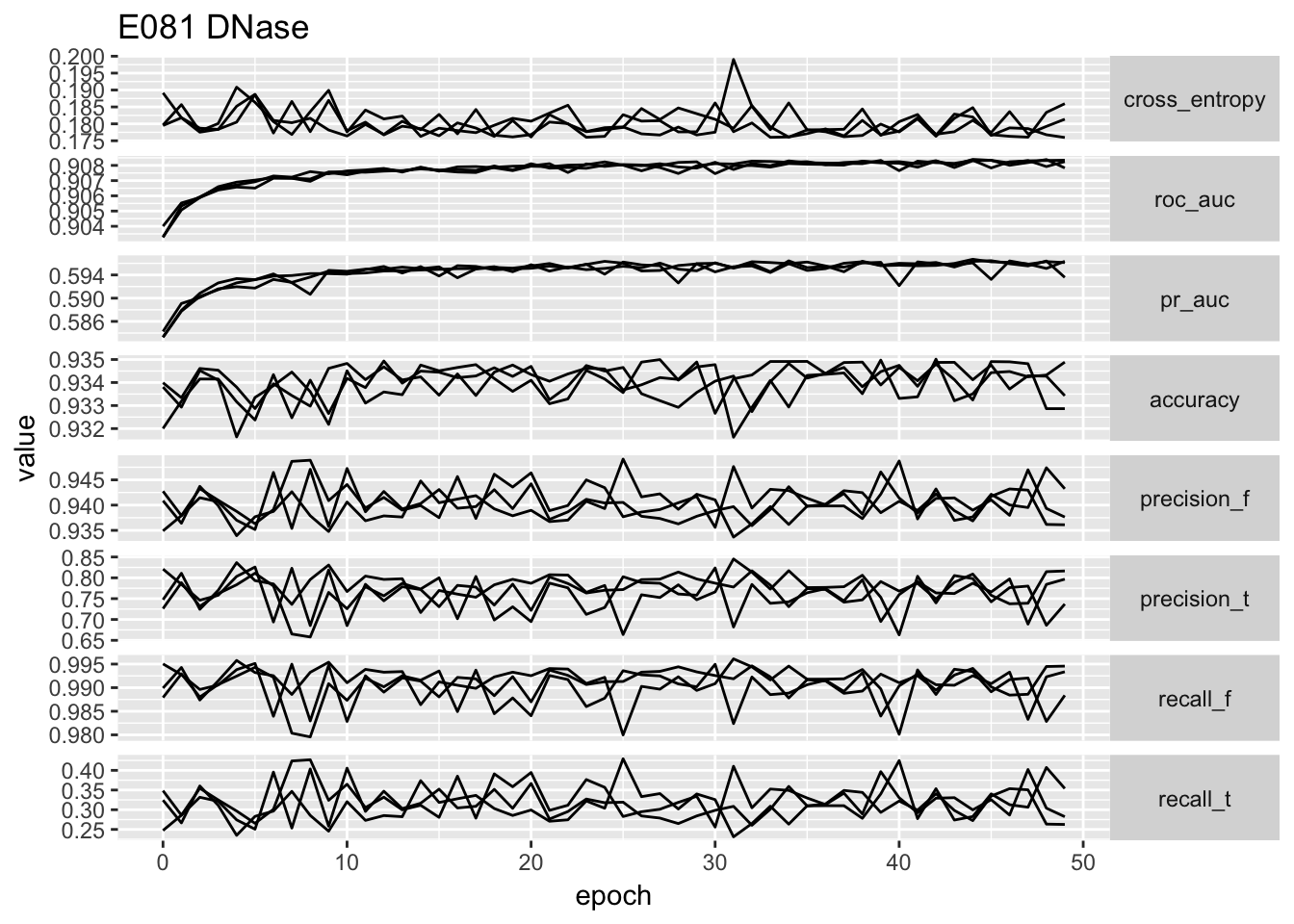
ggplot(temp.no_proportion[temp.no_proportion$data == 'E082',]) + geom_line(aes(x = epoch, y = value, group = repeatID)) + facet_grid(type ~ ., scales = 'free_y', ) + ggtitle('E082 DNase') + theme(strip.text.y = element_text(angle = 0))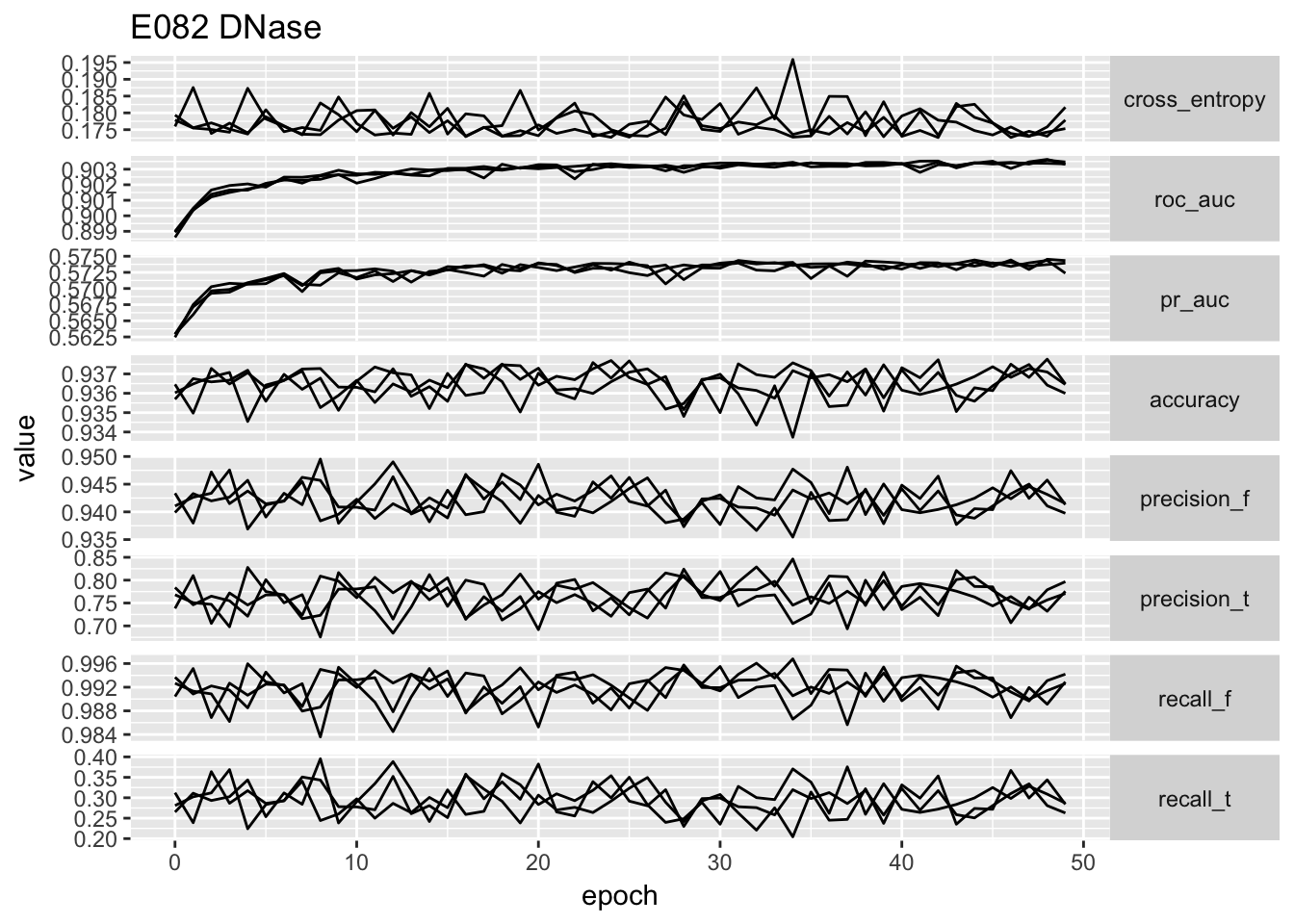
# ggplot(temp) + geom_point(aes(x = cross_entropy, y = roc_auc)) + facet_wrap(~data, scales = 'free')From the performance curves we can see that the present feature representation works well on E081, E082 DNase data but it works really poorly on Noonan’s H3K27ac data. First of all, the bad performance is hardly due to optimization issue, because we have run 50 epochs and other data sets have reached a reasonable performance. We select the one with lowest validation loss as the best model for down stream analysis.
best_model <- summary_table[summary_table$variable == 'val_loss', ][order(summary_table[summary_table$variable == 'val_loss', ]$value, decreasing = F)[1],]
print(best_model)## X data epoch value variable repeatID
## 4075 0 all 42 0.1703 val_loss 2temp_dcast <- dcast(temp, epoch + repeatID + data ~ type)
best_model_melted <- temp_dcast[temp_dcast$epoch == best_model$epoch & temp_dcast$repeatID == best_model$repeatID,]
write.table(best_model_melted, file='../data/best_model_type1_scores.txt', sep = '\t', row.names = F, quote = F)4.4 Compare performance of best model with DeepSEA result
source('~/Box Sync/2017_spring/tasks/deep_variant/DeepSEA/doc/cell_type/yanyu_lib.R')
library(stringr)
aucs_danq <- read.table('../../DeepVariantAnnotation/data/aucs.txt', sep = '\t', header = T)
aucs_danq <- as_num(aucs_danq)## Warning in as_num(aucs_danq): NAs introduced by coercion
## Warning in as_num(aucs_danq): NAs introduced by coercionaucs_danq <- aucs_danq[!is.na(aucs_danq$DeepSEA.ROC.AUC),]
aucs_danq$AnnotationType <- to_annotation_type(aucs_danq$TF.DNase.HistoneMark)
temp_data <- aucs_danq[aucs_danq$AnnotationType == 'DNase',]
p1<-ggplot(aucs_danq) + geom_histogram(aes(x = DeepSEA.ROC.AUC, fill = AnnotationType), bins=50) + facet_grid(AnnotationType~., scales = 'free_y') + theme(strip.background = element_blank(), strip.text.y = element_blank(),legend.position="none") + scale_x_continuous(limits=c(0.25, 1))
p2<-ggplot(aucs_danq) + geom_histogram(aes(x = DeepSEA.PR.AUC, fill = AnnotationType), bins=30) + facet_grid(.~AnnotationType, scales = 'free_y') + coord_flip() + theme(strip.background = element_blank(), strip.text.x = element_blank(), legend.position="none")
p3<-ggplot() + geom_point(data = aucs_danq, aes(x = DeepSEA.ROC.AUC, y = DeepSEA.PR.AUC, color = AnnotationType)) + theme( legend.box = "horizontal", legend.justification=c(0,1), legend.position=c(0,1), legend.text=element_text(size=5), legend.title=element_text(size=5)) + geom_point(data = best_model_melted, aes(x = roc_auc, y = pr_auc, shape = data)) + scale_x_continuous(limits=c(0.25, 1))
multiplot(p3, p1, p2, cols=2)## Warning: Removed 3 rows containing missing values (geom_bar).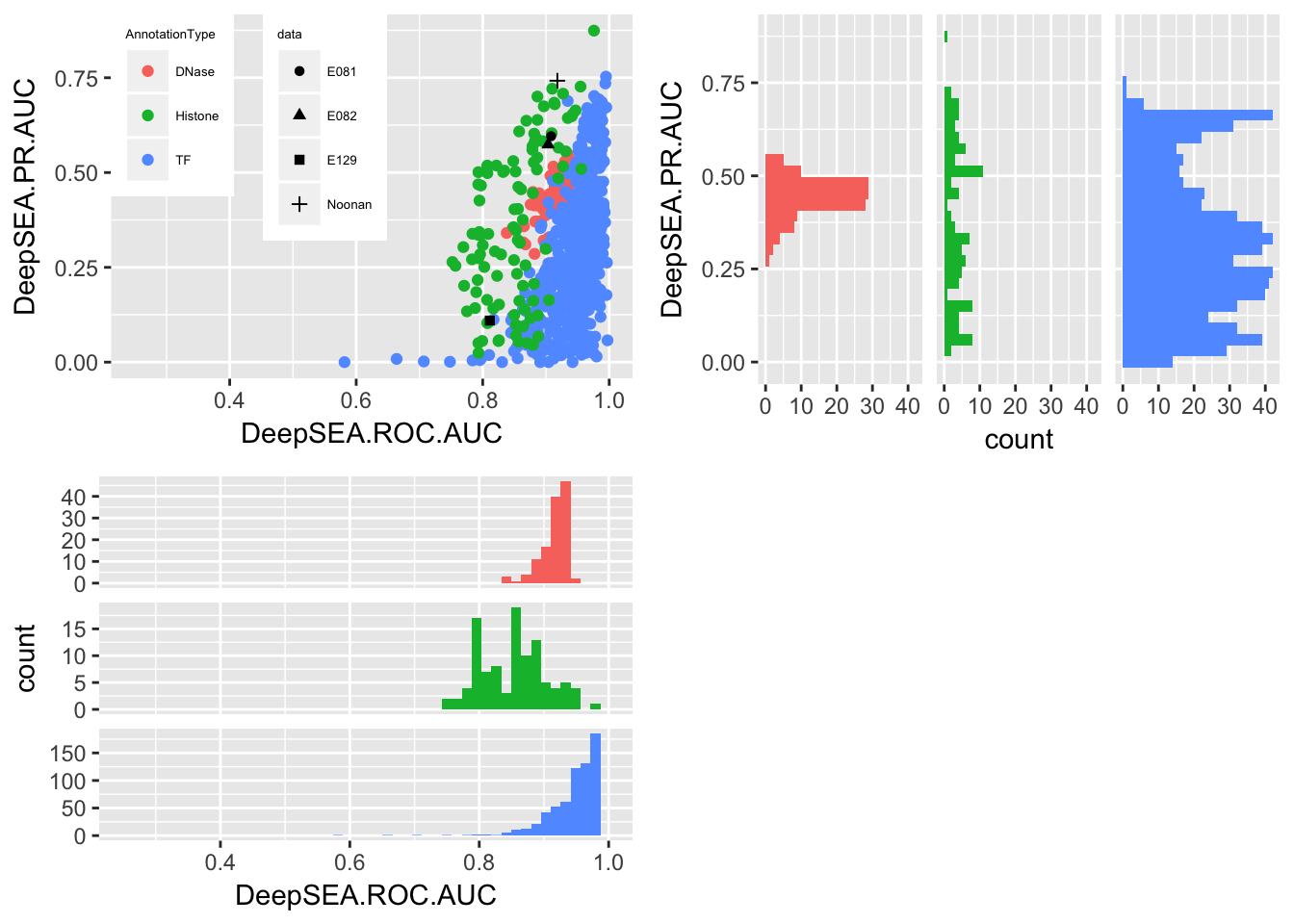
It shows that the result of E081, E082 are compatible to the DNase result in DeepSEA. E129 which is one of the data set in DeepSEA, it achieves similar result as DeepSEA one. But the H3K27ac data behaves poorly, which indicates that the feature representation is not suitable for this data. Another issue is that it is possible that the poor performance data sets have a more unbalanced label set. TODO show that!