Deep brain - Type 1 - evaluation on allelic imbalance instances
07 June, 2017
1 About alignment bias
It has not been implemented. TODO
The basic steps to deal with alignment bias is as follow:
- Align and call all possible heterozygous site
- Use
WASPto look for reads at possible heterozygous site - Re-align those reads and filter out the ones that cannot be aligned to the same position after flip the base
- Call heterozygous site with quality control and compute the degree of imbalance
2 Results
2.1 Load Data
source('my_r.R')
library(feather)
library(ggplot2)
library(reshape2)
variant_url <- 'https://github.com/liangyy/deep_variant/blob/code/allelic_imbalance/data/E081.combined.imbalance.gz?raw=true'
variant <- read.table(read_gz_url(variant_url), sep = '\t', header = T)
score_url <- 'https://raw.githubusercontent.com/CreRecombinase/DeepVariantPrediction/issue1/score/keras_deepsea_with_040417_2_42_head_copy/allelic_imbalance_E081.E081_allelic_imbalance_result.feather'
score <- read_feather_url(score_url)## Warning: Coercing int64 to doublevariant$Ref <- NA
variant$Ref[score$Varient.ID] <- score$Allele1
variant$Alt <- NA
variant$Alt[score$Varient.ID] <- score$Allele22.2 Call Imbalance
variant$af <- variant$Reads1 / (variant$Reads1 + variant$Reads2)
variant <- variant[variant$af < 0.8,]
imbalance <- ( variant$af > 0.7 | variant$af < 0.4 ) & variant$P.Value < 0.01
balance <- 0.45 < variant$af & variant$af < 0.55 & variant$P.Value > 0.05
# imbalance.threshold <- 0.01 # Fisher's exact test p-value
variant$Imbalance <- 'Unsure'
variant$Imbalance[imbalance] <- 'Imbalanced'
variant$Imbalance[balance] <- 'Balanced'
ggplot(variant[variant$Imbalance == 'Imbalanced', ]) + geom_bar(aes(x = af > 0.5)) + ggtitle('Reference Biased vs Alternative Biased') + xlab('Reference Biased')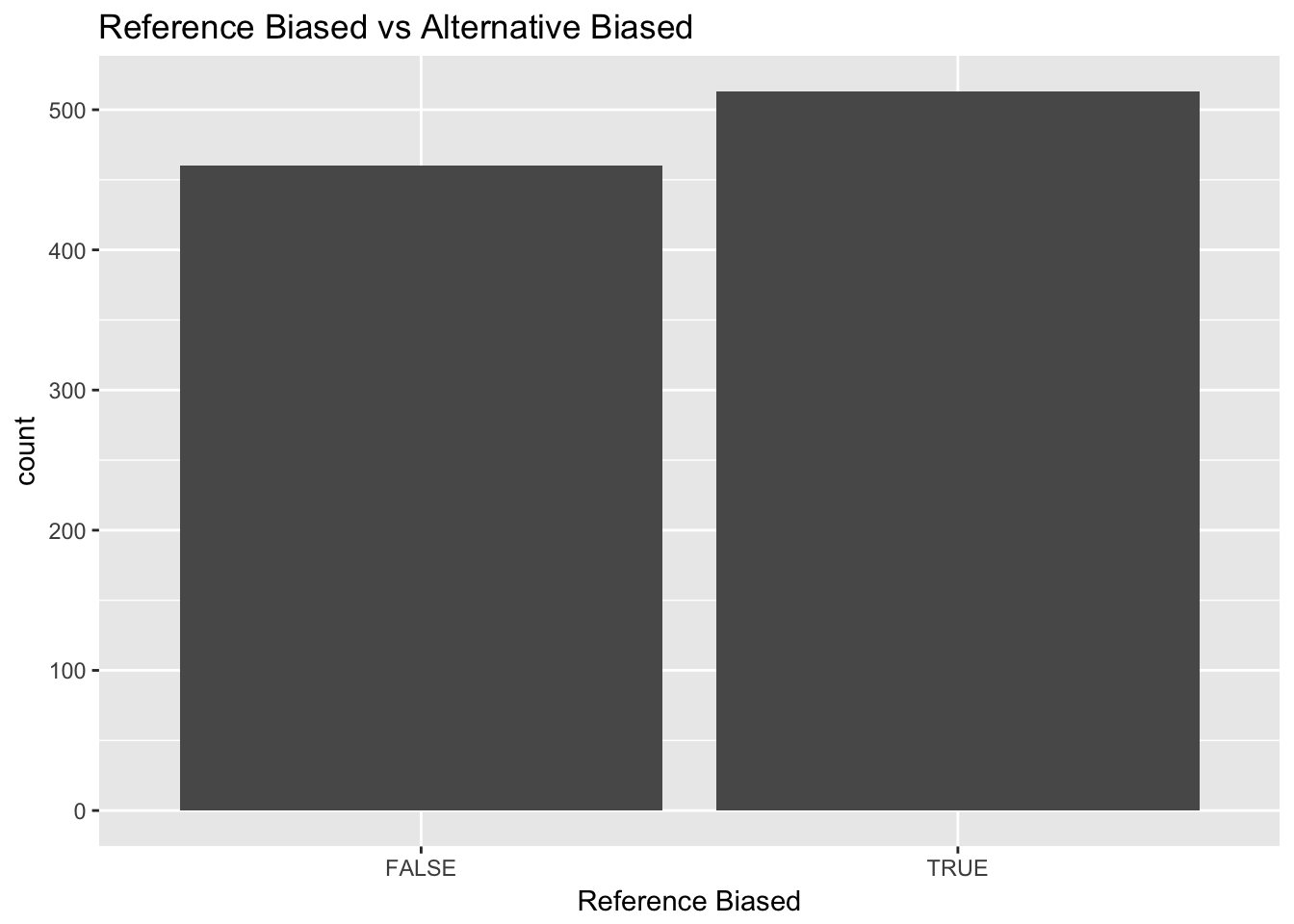
2.3 Predicted imbalance
2.3.1 Overview
ggplot(variant) + geom_point(aes(x = Alt, y = Ref, color = log10(Odds.Ratio))) + facet_grid(.~Imbalance) + geom_abline(intercept = 0.07, slope = 1) + geom_abline(intercept = -0.07, slope = 1) + ggtitle('The Predicted Effect of \n Heterozygous sites in DNase-seq of E081') + scale_color_gradient2(midpoint=0, low="blue", mid="white", high="red", space ="Lab" )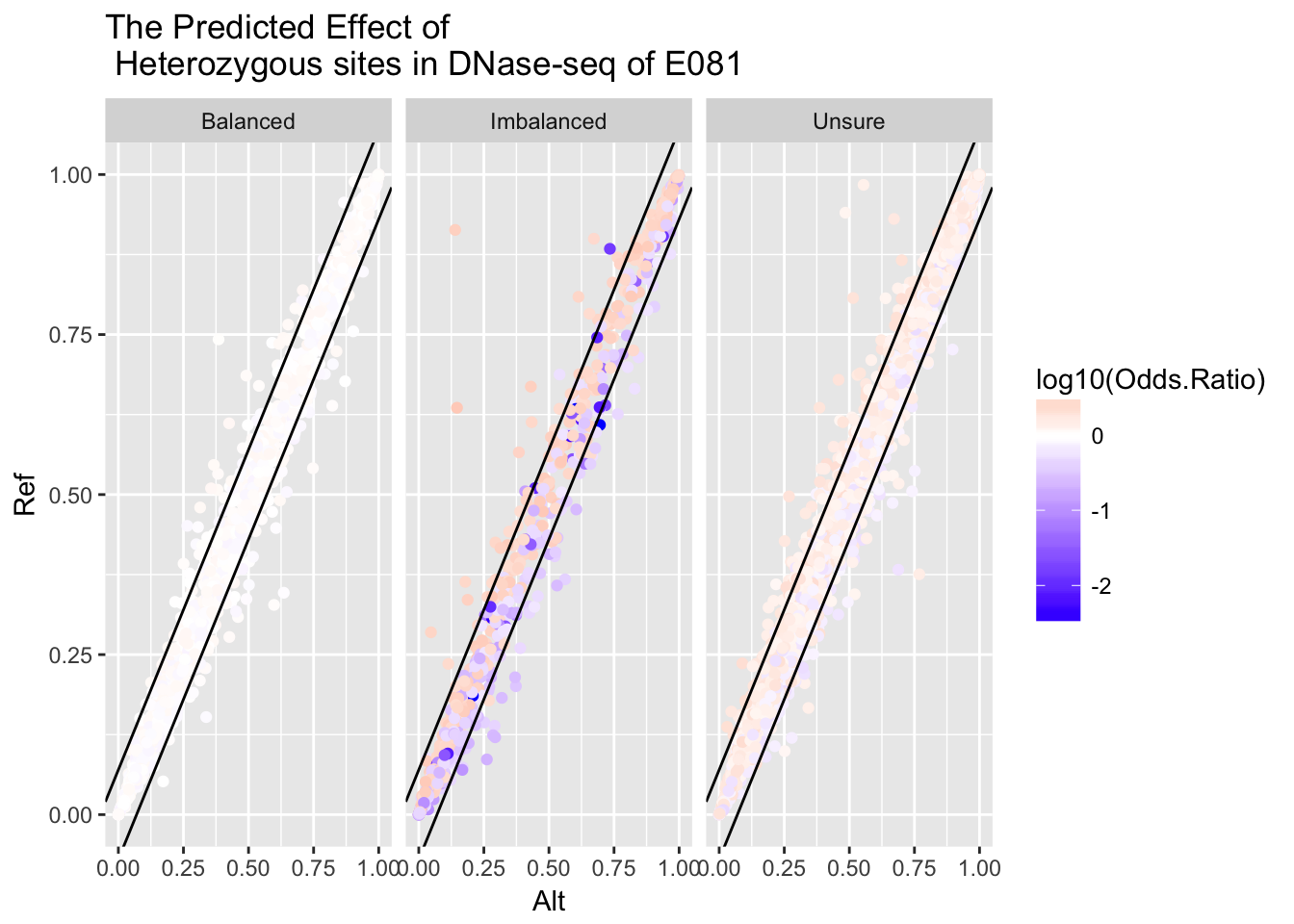
2.3.2 Observed Log Odds Ratio vs. Predicted Log Odds Ratio
variant$logit.raf <- logit(variant$af)
variant$log10.oddsratio.predicted <- logit(variant$Ref) - logit(variant$Alt)
variant$logit.raf.predicted <- logit(variant$Ref / (variant$Ref + variant$Alt))
model.linear = lm(logit.raf ~ 0 + logit.raf.predicted, data = variant)
ggplot(variant, aes(x = logit.raf.predicted, y = logit.raf)) + geom_point(aes(color = Imbalance)) + geom_smooth(method='lm',formula=y~0+x) + annotate("text", x = 0.4, y = -1, label = lm_eqn(model.linear), parse = TRUE) + ggtitle('Observed Reference Allele Frequency vs \n Predicted RAF')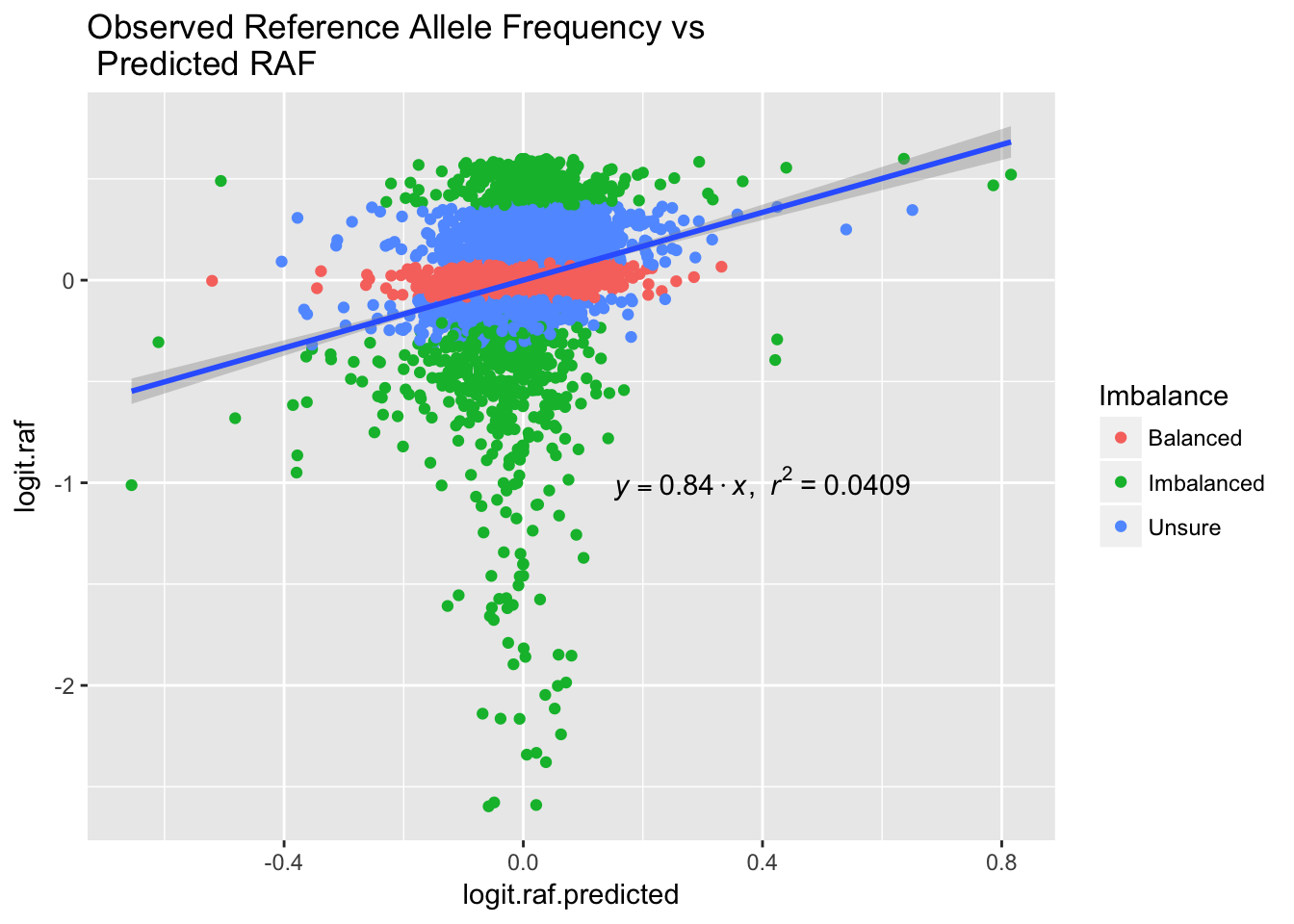
2.3.3 Predictive power of predicted p(ref) and p(alt)
Model: imbalance ~ f(ref, alt), link = binomial. \(f_1 = |x - y|\). \(f_2 = |\log_{10}(x/(1-x)) - \log_{10}(y/(1-y))|\). \(f_3 = logit(\frac{p_0}{p_0 + p_1}) = \log_{10} \frac{p_0}{p_1}\)
variant.pass.interest <- variant[variant$Imbalance != 'Unsure',]
model.logistic.abs <- glm(as.numeric(Imbalance == 'Imbalanced') ~ abs(Ref - Alt), family=binomial(link='logit'), data=variant.pass.interest)
model.logistic.lor <- glm(as.numeric(Imbalance == 'Imbalanced') ~ abs(log10.oddsratio.predicted), family=binomial(link='logit'), data=variant.pass.interest)
model.logistic.logitraf <- glm(as.numeric(Imbalance == 'Imbalanced') ~ abs(logit.raf.predicted), family=binomial(link='logit'), data=variant.pass.interest)library(pander)
panderOptions('knitr.auto.asis', FALSE)
pander(model.logistic.abs)| Estimate | Std. Error | z value | Pr(>|z|) | |
|---|---|---|---|---|
| abs(Ref - Alt) | 6.96 | 1.179 | 5.902 | 3.599e-09 |
| (Intercept) | -1.056 | 0.04628 | -22.83 | 2.401e-115 |
pander(model.logistic.lor)| Estimate | Std. Error | z value | Pr(>|z|) | |
|---|---|---|---|---|
| abs(log10.oddsratio.predicted) | 3.099 | 0.4461 | 6.947 | 3.722e-12 |
| (Intercept) | -1.166 | 0.05408 | -21.56 | 4.026e-103 |
pander(model.logistic.logitraf)| Estimate | Std. Error | z value | Pr(>|z|) | |
|---|---|---|---|---|
| abs(logit.raf.predicted) | 9.13 | 0.7761 | 11.76 | 5.975e-32 |
| (Intercept) | -1.265 | 0.04968 | -25.46 | 6.049e-143 |
library(precrec)
joined <- join_scores(model.logistic.abs$fitted.values, model.logistic.lor$fitted.values, model.logistic.logitraf$fitted.values)
msmdat <- mmdata(joined, as.numeric(variant.pass.interest$Imbalance == 'Imbalanced'), modnames = c('f1', 'f2', 'f3'))
mscurves <- evalmod(msmdat)
plot(mscurves)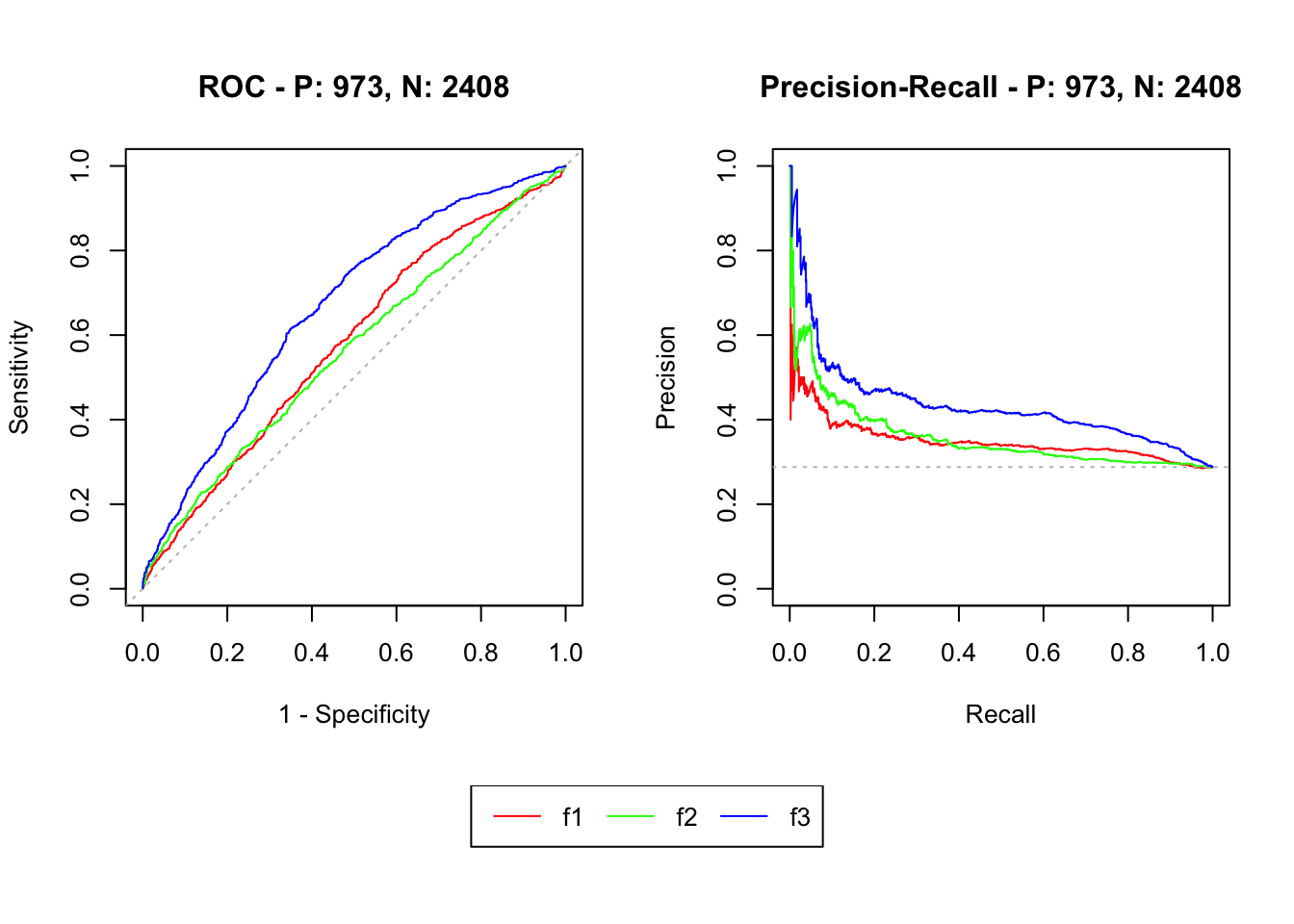
pander(auc(mscurves))| modnames | dsids | curvetypes | aucs |
|---|---|---|---|
| f1 | 1 | ROC | 0.58 |
| f1 | 1 | PRC | 0.3507 |
| f2 | 1 | ROC | 0.5636 |
| f2 | 1 | PRC | 0.3586 |
| f3 | 1 | ROC | 0.6673 |
| f3 | 1 | PRC | 0.435 |